Neuronal RNAs are made in the cell body and transported into axons and dendrites – how do these RNAs find their way to their final destination? Excited about our Nature Neuroscience paper, a joint effort with Igor Ulitsky’s lab, in which we describe a method for massively parallel mapping of mRNA localization elements and identify the let-7 binding site and (AU)n motif as novel localization elements in primary cortical neurons.
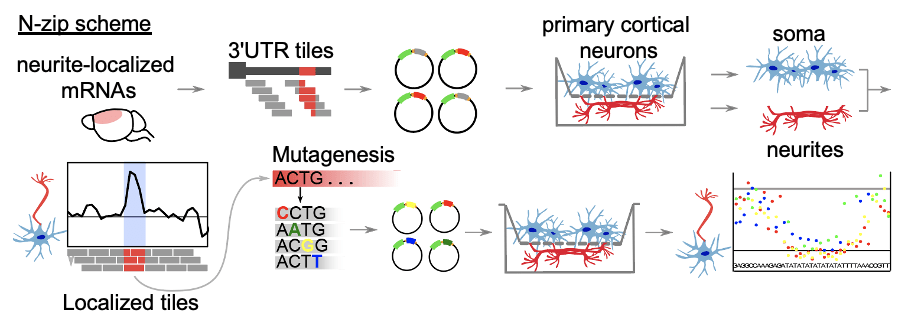
The localization of mRNAs is thought to depend largely on specific sequences present in mRNAs – “zipcodes” – which are bound by RNA-binding proteins (RBPs) that direct them to their sites of function. Hundreds to thousands of mRNAs are known to be localized in neurons, but the zipcodes and RBPs that bind to them have been identified only for a few. Furthermore, it hasn’t been known if binding to noncoding RNAs plays a role in sculpting mRNA localization. Here, we tackle this problem by integrating our previously established methods to separate the subcellular compartments of neurons – soma and neurites – for omics analyses (Zappulo et al. Nature Comm 2017) and then use massively parallel reporter assay (MPRA) for the mapping of RNA nuclear localization signals (Lubelsky and Ulitsky, Nature 2018). The new methodology allows for an unbiased, transcriptome-wide mapping of zipcodes, which we call neuronal zipcode identification protocol (N-zip).
Applying N-zip to primary cortical neurons allowed us to identify two de novo zipcodes: a binding site for let-7 miRNAs and an (AU)n motif, and RBPs bound to them. We have also identified the mechanism by which these sequences regulate localization to neurites, through a preferential degradation of transcripts in the soma. The process we describe represents a novel mechanism that links miRNA activity to neuronal mRNA localization.
The N-zip technique can be extended to diverse types of neurons to observe molecular processes in contexts such as healthy development, learning and neurological diseases. It can also be used to study other polarized cell types such as fibroblasts and cancer cells. It’s an important step toward unraveling how a finite number of patterns in cells produce many of highly intricate polarized cell types and help them adapt to changing environments in health and disease.
Congratulations to Samantha, Nicolai, Sayaka, Maya and other co-authors!
https://www.nature.com/articles/s41593-022-01243-x